Label-free HSI for Surgical Margin Assessment
Quantification Tools for HSI During Surgical Guidance
Spectral-Spatial Classification Methods for Tumor Detection
Minimum Spanning Forest Based Method for HSI
The objective is to develop a classification method that combines both spectral and spatial information for distinguishing cancer from healthy tissue on hyperspectral images in an animal model. An automated algorithm based on a minimum spanning forest (MSF) and optimal band selection has been proposed to classify healthy and cancerous tissue on hyperspectral images. A support vector machine (SVM) classifier is trained to create a pixel-wise classification probability map of cancerous and healthy tissue. This map is then used to identify markers that are used to compute mutual information for a range of bands in the hyperspectral image and thus select the optimal bands. An MSF is finally grown to segment the image using spatial and spectral information. The MSF based method with automatically selected bands proved to be accurate in determining the tumor boundary on hyperspectral images. Hyperspectral imaging combined with the proposed classification technique has the potential to provide a noninvasive tool for cancer detection.
MSF Process
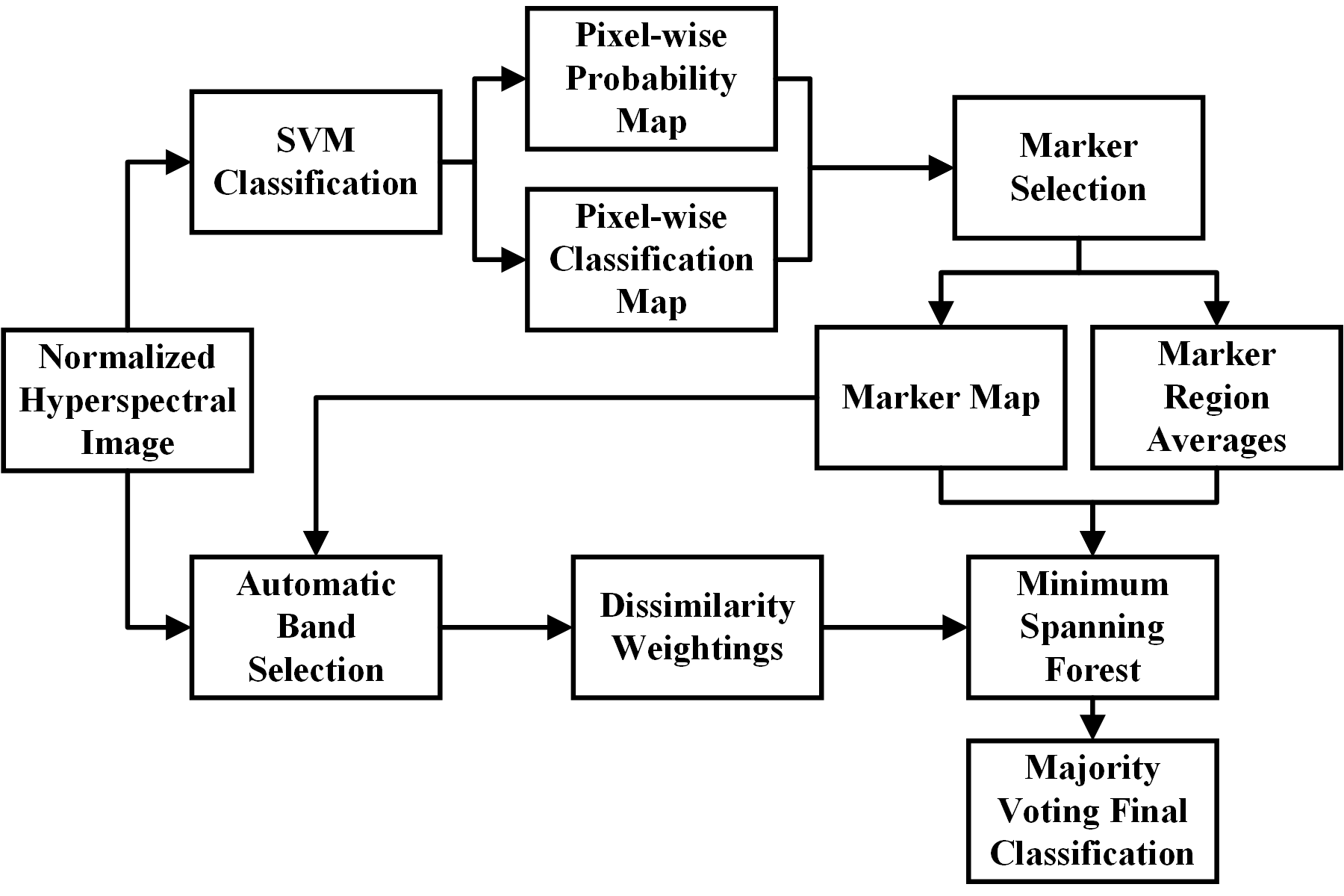
Flowchart of the minimum spanning forest based method for the classification of normal and cancer tissue.
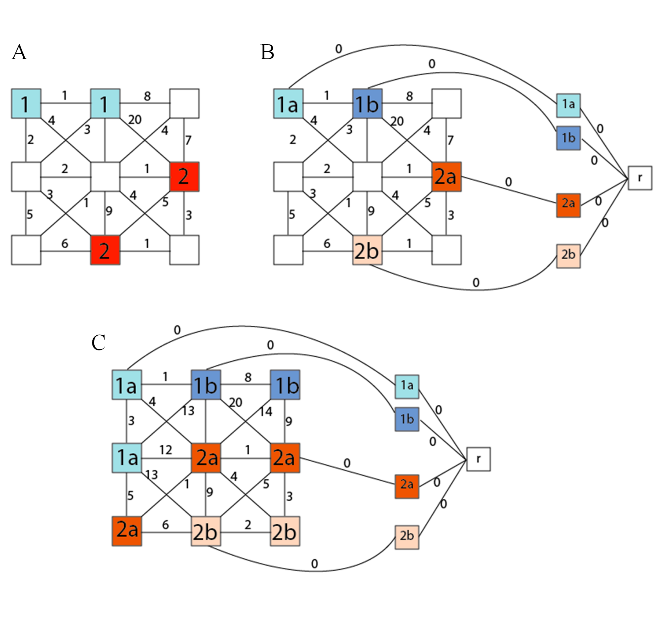
Flow Visualization of the MSF construction from the marker selection (A) to the marker labeling (B) and to the complete construction of the MSF (C) that will be classified with majority voting.
Mouse Spectral Result
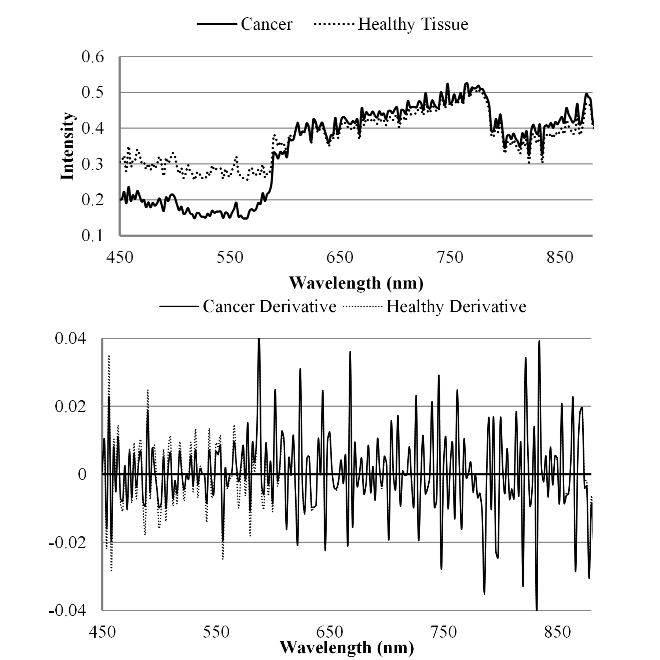
Spectral curves of the mouse image and the derivative of the image for both cancerous and healthy tissue.